Volume 22, Issue 5 (11-2019)
J Arak Uni Med Sci 2019, 22(5): 68-77 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Fallahzadeh R, Esfahani K, Akhavan Sepahi A, Kamali N, Bambai B. Increasing the catalytic power of the flavin reductase DszD enzyme using site-directed mutagenesis method in Rhodococcus erythropolis. J Arak Uni Med Sci 2019; 22 (5) :68-77
URL: http://jams.arakmu.ac.ir/article-1-6094-en.html
URL: http://jams.arakmu.ac.ir/article-1-6094-en.html
1- Department of Microbiology, Tehran North Branch of Islamic Azad University, Tehran, Iran.
2- Department of Medical Genetics, Institute of Medical Biotechnology, National Institute of Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran.
3- Department of Medical Genetics, Institute of Medical Biotechnology, National Institute of Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran. ,bambai@nigeb.ac.ir
2- Department of Medical Genetics, Institute of Medical Biotechnology, National Institute of Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran.
3- Department of Medical Genetics, Institute of Medical Biotechnology, National Institute of Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran. ,
Full-Text [PDF 2730 kb]
(1455 Downloads)
| Abstract (HTML) (3991 Views)
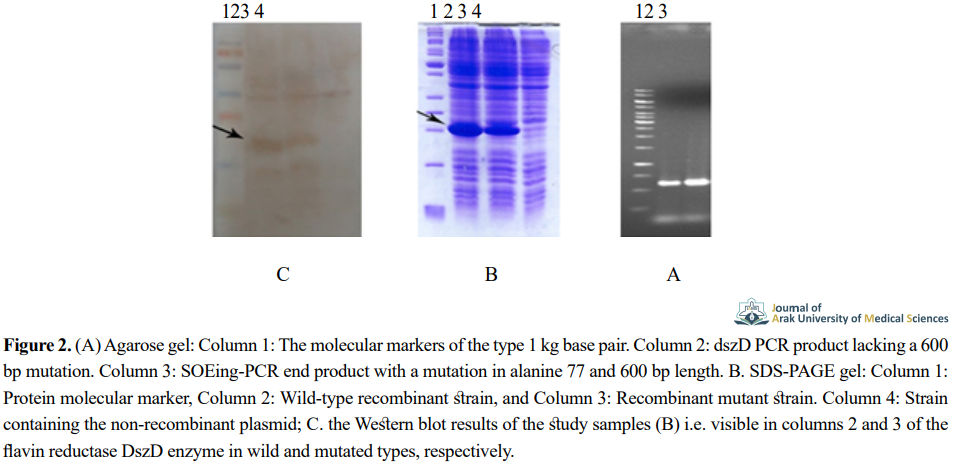
Wild-type dszD gene amplification was performed applying specific primers (Figure 2a, Column 1). The mutated dszD gene was also obtained at position 77 by a mutant primer (Figure 2a, Column 2). After the enzymatic digestion of the pET-23a(+) vector and the final amplified fragments, each was separately ligated to generate the recombinant plasmid during the binding reaction.
The recombinant plasmids containing the wild- and mutated-type genes were individually transferred to the E.coli BL21 (DE3) expression strain. The experimental results indicated the appropriate expression of the target cloned genes; it had a specific band on the polyacrylamide gel with a molecular weight of 24 kDa (Figure 2b).
Western blotting analysis supported the presence of a significant level of expression of the wild and mutated types of recombinant flavin reductase DszD enzyme (Figure 2c). We compared the mean catalytic power of the wild-type and mutant-type flavin reductase DszD. The obtained data revealed that the mutant enzyme had a catalytic capacity of 392±6 U/mg, suggesting a 2.5-fold increase in activity, compared to the wild-type enzyme (160±4 U/mg).
Conclusion
The achieved data suggested that the target enzyme had an appropriate potential to increase the catalytic power in the 4S route. Thus, the targeted mutation in the flavin reductase DszD enzyme and its activity in recombinant host cells could increase the desulfurization process efficacy, using the bacterial system. It is suggested that different mutant molecules capable of mutation be produced in several key positions, and their catalytic potency be compared with those reported.
Ethical Considerations
Compliance with ethical guidelines
This study with research ethics code IR.NIGEB.EC.1398.6.24 A has been approved by research ethics committee at National Institute of Genetic Engineering and Biotechnology, Tehran, Iran.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Authors' contributions
Conceptualization, methodology, investigation, writing-original draft, writing-review & editing: all author.
Conflicts of interest
The authors declared no conflict of interest.
References
Full-Text: (2421 Views)
Introduction
The combustion of petroleum compounds, due to the release of sulfur oxides, can cause numerous environmental issues, like air pollution, as well as many cardiac, respiratory, dermatological, and gastrointestinal diseases in individuals. Various approaches are applied to reduce the sulfur content of petroleum compounds in the petroleum refinery; chemical desulfurization is the most frequently used method in this respect. The chemical desulfurization is inefficient and requires powerful thermal systems, high pressure, high energy consumption, and costly metal catalysts. The microbial desulfurization method has recently been developed. The microbial desulfurization could remove sulfur in the polycyclic derivatives of petroleum without breaking the carbon skeleton and reducing fuel energy value along the 4S route. This pathway was first introduced in the gram-negative Rhodococcus erythropolis bacteria (Rhodococcus erythropolis IGTS8). This bacterium uses sulfur in dibenzothiophene and its derivatives as the sole nutrition source. The active desulfurization reaction is performed in the presence of the flavin reductase DszD enzyme. Naturally, this enzyme has low catalytic power; therefore, it is considered as the primary limiting factor in the desulfurization process rate in the refining industry. The current study aimed to increase the catalytic power of the target enzyme. Thus, the asparagine residue at position 77 was replaced with phenylaniline by targeted mutagenesis. Eventually, the catalytic power of wild and mutated enzymes was compared.
Materials and methods
The DszD enzyme amino acid sequence is accessible at the National Center for Biotechnology Information (NCBI) database under the code AAC38226.1. To identify homologous molecules with acceptable similarity, the target enzyme sequence was searched in the protein database. Among the identified homologs, those bounded to the FMN substrate were selected, and their alignment was performed using the Basic Local Alignment Search Tool (BLAST). The wild-type dszD gene was amplified by Polymerase Chain Reaction (PCR) using forward and backward primers with cleavage sites for BamHI and EcoRI restriction enzymes, respectively. The dszD gene was mutated at position 77 using SOEing-PCR. The mutated wild-type gene of 600 bp length was individually cloned into the pET-23a(+) expression vector, then transferred to E. coli BL21 (DE3) cells. Cloning accuracy was assessed using the two restriction enzymes mentioned above and sequencing. Wild and mutated proteins were separately expressed and confirmed by the Western blotting technique. Next, their optical absorption was measured at 340 nm during the oxidation of NADH to NAD per time unit. Protein concentration was determined by the Bradford method with Bovine Serum Albumin (BSA) as the standard protocol.
Results
Searching for the amino acid sequence of the flavin reductase DszD enzyme in the protein database has introduced homologous molecules with distinct crystallographic structure. Homologous molecules with the highest structural similarity were selected as appropriate models for identifying key positions. The alignment of the amino acid sequence of DszD enzyme with the selected homologous molecules revealed that the target enzyme active site includes 4 key positions for binding to the FMN substrate; threonine 62, serine 63, asparagine 77, and alanine 79 (Figure 1).
The combustion of petroleum compounds, due to the release of sulfur oxides, can cause numerous environmental issues, like air pollution, as well as many cardiac, respiratory, dermatological, and gastrointestinal diseases in individuals. Various approaches are applied to reduce the sulfur content of petroleum compounds in the petroleum refinery; chemical desulfurization is the most frequently used method in this respect. The chemical desulfurization is inefficient and requires powerful thermal systems, high pressure, high energy consumption, and costly metal catalysts. The microbial desulfurization method has recently been developed. The microbial desulfurization could remove sulfur in the polycyclic derivatives of petroleum without breaking the carbon skeleton and reducing fuel energy value along the 4S route. This pathway was first introduced in the gram-negative Rhodococcus erythropolis bacteria (Rhodococcus erythropolis IGTS8). This bacterium uses sulfur in dibenzothiophene and its derivatives as the sole nutrition source. The active desulfurization reaction is performed in the presence of the flavin reductase DszD enzyme. Naturally, this enzyme has low catalytic power; therefore, it is considered as the primary limiting factor in the desulfurization process rate in the refining industry. The current study aimed to increase the catalytic power of the target enzyme. Thus, the asparagine residue at position 77 was replaced with phenylaniline by targeted mutagenesis. Eventually, the catalytic power of wild and mutated enzymes was compared.
Materials and methods
The DszD enzyme amino acid sequence is accessible at the National Center for Biotechnology Information (NCBI) database under the code AAC38226.1. To identify homologous molecules with acceptable similarity, the target enzyme sequence was searched in the protein database. Among the identified homologs, those bounded to the FMN substrate were selected, and their alignment was performed using the Basic Local Alignment Search Tool (BLAST). The wild-type dszD gene was amplified by Polymerase Chain Reaction (PCR) using forward and backward primers with cleavage sites for BamHI and EcoRI restriction enzymes, respectively. The dszD gene was mutated at position 77 using SOEing-PCR. The mutated wild-type gene of 600 bp length was individually cloned into the pET-23a(+) expression vector, then transferred to E. coli BL21 (DE3) cells. Cloning accuracy was assessed using the two restriction enzymes mentioned above and sequencing. Wild and mutated proteins were separately expressed and confirmed by the Western blotting technique. Next, their optical absorption was measured at 340 nm during the oxidation of NADH to NAD per time unit. Protein concentration was determined by the Bradford method with Bovine Serum Albumin (BSA) as the standard protocol.
Results
Searching for the amino acid sequence of the flavin reductase DszD enzyme in the protein database has introduced homologous molecules with distinct crystallographic structure. Homologous molecules with the highest structural similarity were selected as appropriate models for identifying key positions. The alignment of the amino acid sequence of DszD enzyme with the selected homologous molecules revealed that the target enzyme active site includes 4 key positions for binding to the FMN substrate; threonine 62, serine 63, asparagine 77, and alanine 79 (Figure 1).
Wild-type dszD gene amplification was performed applying specific primers (Figure 2a, Column 1). The mutated dszD gene was also obtained at position 77 by a mutant primer (Figure 2a, Column 2). After the enzymatic digestion of the pET-23a(+) vector and the final amplified fragments, each was separately ligated to generate the recombinant plasmid during the binding reaction.
The recombinant plasmids containing the wild- and mutated-type genes were individually transferred to the E.coli BL21 (DE3) expression strain. The experimental results indicated the appropriate expression of the target cloned genes; it had a specific band on the polyacrylamide gel with a molecular weight of 24 kDa (Figure 2b).
Western blotting analysis supported the presence of a significant level of expression of the wild and mutated types of recombinant flavin reductase DszD enzyme (Figure 2c). We compared the mean catalytic power of the wild-type and mutant-type flavin reductase DszD. The obtained data revealed that the mutant enzyme had a catalytic capacity of 392±6 U/mg, suggesting a 2.5-fold increase in activity, compared to the wild-type enzyme (160±4 U/mg).
Conclusion
The achieved data suggested that the target enzyme had an appropriate potential to increase the catalytic power in the 4S route. Thus, the targeted mutation in the flavin reductase DszD enzyme and its activity in recombinant host cells could increase the desulfurization process efficacy, using the bacterial system. It is suggested that different mutant molecules capable of mutation be produced in several key positions, and their catalytic potency be compared with those reported.
Ethical Considerations
Compliance with ethical guidelines
This study with research ethics code IR.NIGEB.EC.1398.6.24 A has been approved by research ethics committee at National Institute of Genetic Engineering and Biotechnology, Tehran, Iran.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Authors' contributions
Conceptualization, methodology, investigation, writing-original draft, writing-review & editing: all author.
Conflicts of interest
The authors declared no conflict of interest.
References
- Etemadi N, Sepahy AA, Mohebali G, Yazdian F, Omidi M. Enhancement of bio-desulfurization capability of a newly isolated thermophilic bacterium using starch/iron nanoparticles in a controlled system. Int J biol macromol. 2018; 120:1801-9. [DOI:10.1016/j.ijbiomac.2018.09.110] [PMID]
- Kilbane II JJ. Microbial biocatalyst developments to upgrade fossil fuels. Curr Opin Biotechnol. 2006; 17(3):305-14. [DOI:10.1016/j.copbio.2006.04.005] [PMID]
- Chen S, Zhao C, Liu Q, Zang M, Liu C, Zhang Y. Thermophilic biodesulfurization and its application in oil desulfurization. Appl Microbiol Biotechnol. 2018; 102(21):9089-103. [DOI:10.1007/s00253-018-9342-5] [PMID]
- Nuhu AA. Bio-catalytic desulfurization of fossil fuels: A mini review. Rev in Envir Sci Bio/Technol. 2013; 12(1):9-23. [DOI:10.1007/s11157-012-9267-x]
- Morrison E, Kantz A, Gassner GT, Sazinsky MH. Structure and mechanism of styrene monooxygenase reductase: New insight into the FAD-transfer reaction. Biochem. 2013; 52(35):6063-75. [DOI:10.1021/bi400763h] [PMID] [PMCID]
- Gupta N, Roychoudhury P, Deb J. Biotechnology of desulfurization of diesel: Prospects and challenges. Appl Microbiol Biotechnol. 2005; 66(4):356-66. [DOI:10.1007/s00253-004-1755-7] [PMID]
- Davoodi-Dehaghani F, Vosoughi M, Ziaee AA. Biodesulfurization of dibenzothiophene by a newly isolated Rhodococcus erythropolis strain. Bioresour Technol. 2010; 101(3):1102-5. [DOI:10.1016/j.biortech.2009.08.058] [PMID]
- Zhang M, Zhu W, Xun S, Li H, Gu Q, Zhao Z, et al. Deep oxidative desulfurization of dibenzothiophene with POM-based hybrid materials in ionic liquids. Chem engineer J. 2013; 220:328-36. [DOI:10.1016/j.cej.2012.11.138]
- Xiao J, Wang X, Fujii M, Yang Q, Song C. A novel approach for ultra‐deep adsorptive desulfurization of diesel fuel over TiO2-CeO2/MCM‐48 under ambient conditions. AIChE J. 2013; 59(5):1441-5. [DOI:10.1002/aic.14085]
- Borzenkova N, Veselova I, Shekhovtsova T. Biochemical methods of crude hydrocarbon desulfurization. Biol Bull Rev. 2013; 3(4):296-311. [DOI:10.1134/S2079086413040026]
- Sucharitakul J, Tinikul R, Chaiyen P. Mechanisms of reduced flavin transfer in the two-component flavin-dependent monooxygenases. Arch biochem biophysics. 2014; 555-556:33-46. [DOI:10.1016/j.abb.2014.05.009] [PMID]
- Nazari F, Kefayati M, Raheb J. The study of biological technologies for the removal of sulfur compounds. J Sci, Islam Repub Iran. 2017; 28(3):205-19.
- Karimi E, Yazdian F, Rasekh B, Jeffryes C, Rashedi H, Akhavan Sepah A, et al. DBT desulfurization by decorating bacteria using modified carbon nanotube. Fuel. 2018; 216:787-95. [DOI:10.1016/j.fuel.2017.10.030]
- Khosravinia S, Mahdavi MA, Gheshlaghi R, Dehghani H, Rasekh B. Construction and characterization of a new recombinant vector to remove sulfate repression of dsz promoter transcription in biodesulfurization of dibenzothiophene. Frontiers Microbiol. 2018; 9(1578):1-9. [DOI:10.3389/fmicb.2018.01578] [PMID] [PMCID]
- Martínez I, Mohamed ME-S, Santos VE, García JL, García-Ochoa F, Díaz E. Metabolic and process engineering for biodesulfurization in Gram-negative bacteria. J biotechnol. 2017; 262:47-55. [DOI:10.1016/j.jbiotec.2017.09.004] [PMID]
- Sousa SrF, Sousa JF, Barbosa AC, Ferreira CE, Neves RP, Ribeiro AJ, et al. Improving the biodesulfurization of crude oil and derivatives: A QM/MM investigation of the catalytic mechanism of NADH-FMN oxidoreductase (DszD). J Physical Chem. 2016; 120(27):5300-6. [DOI:10.1021/acs.jpca.6b01536] [PMID]
- Ferreira P, Sousa SF, Fernandes PA, Ramos MJ. Improving the catalytic power of the DszD enzyme for the biodesulfurization of crude oil and derivatives. Chem Eur J. 2017; 23(68):17231-41. [DOI:10.1002/chem.201786864] [PMID]
- Kamali N, Tavallaie M, Bambai B, Karkhane AA, Miri M. Site-directed mutagenesis enhances the activity of NADH-FMN oxidoreductase (DszD) activity of Rhodococcus erythropolis. Biotechnol Letters. 2010; 32(7):921-7. [DOI:10.1007/s10529-010-0254-4] [PMID]
- Magrane M, Consortium U. UniProt knowledgebase: A hub of integrated protein data. Nat Prec. 2010. [DOI:10.1038/npre.2010.5092]
- Young L, Smith HO, Gibson DG. In vitro recombination method. San Diego: Google Patents; 2017.
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989.
- Schägger H. Tricine-sds-page. Nat Protoc. 2006; 1(1):16-22. [DOI:10.1038/nprot.2006.4] [PMID]
- Kruger NJ. The Bradford method for protein quantitation. The protein protocols handbook. Berlin: Springer; 2002. [DOI:10.1385/1-59259-169-8:15]
- Wang J, Butler III RR, Wu F, Pombert JF, Kilbane II JJ, Stark BC. Enhancement of microbial biodesulfurization via genetic engineering and adaptive evolution. PloS One. 2017; 12(1):e0168833. [DOI:10.1371/journal.pone.0168833] [PMID] [PMCID]
- Akhtar N, Ghauri MA, Akhtar K. Dibenzothiophene desulfurization capability and evolutionary divergence of newly isolated bacteria. Arch Microbiol. 2016; 198(6):509-19. [DOI:10.1007/s00203-016-1209-5] [PMID]
- Rangra S, Kabra M, Gupta V, Srivastava P. Improved conversion of Dibenzothiophene into sulfone by surface display of Dibenzothiophene monooxygenase (DszC) in recombinant Escherichia coli. J Biotechnol. 2018; 287:59-67. [DOI:10.1016/j.jbiotec.2018.10.004] [PMID]
- Vollhardt KPC, Schore NE. Organic chemistry; Palgrave version: Structure and function: Macmillan international higher education. New York: W H Freeman; 2014. [DOI:10.1007/978-1-319-19197-9_2]
- Chen H, Li M, Liu C, Zhang H, Xian M, Liu H. Enhancement of the catalytic activity of Isopentenyl Diphosphate Isomerase (IDI) from Saccharomyces cerevisiae through random and site-directed mutagenesis. Microbial Cell Factories. 2018; 17(1):1-14. [DOI:10.1186/s12934-018-0913-z] [PMID] [PMCID]
- Duan X, Chen J, Wu J. Improving the thermostability and catalytic efficiency of Bacillus deramificans pullulanase by site-directed mutagenesis. Appl Environ Microbiol. 2013; 79(13):4072-7. [DOI:10.1128/AEM.00457-13] [PMID] [PMCID]
Type of Study: Original Atricle |
Subject:
Basic Sciences
Received: 2019/06/13 | Accepted: 2019/10/14
Received: 2019/06/13 | Accepted: 2019/10/14
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |